Probing DNA Structure and DNA/Ligand, DNA/Protein Contacts
In order to probe the weak contacts between proteins and nucleic acids in sequence-specific complexes, and to understand the contribution that such contacts make to overall complex stability, isosteric nucleoside analogues will be employed to introduce incremental changes into the DNA recognition site, by what amounts to “atomic mutagenesis” of the functional group character. If a given functional group is critical to the formation of the protein-nucleic acid complex then an incremental response in the ability to form the complex should be present. If a functional group is unimportant, then no change is complex formation should occur. The most useful analogues are those that do not otherwise alter native interstrand hydrogen bonding or other helix-stabilizing effects. The proposal focuses on the functional groups present in the DNA minor groove. A variety of analogue residues are described that maintain normal Watson-Crick hydrogen bonding but alter the nature of the minor groove functional groups. For the pyrimidines, the minor groove O2-carbonyls will be replaced with –H, –F and –CH3. For the purines the N3-nitrogens will be replaced with C-H, or the more sterically intrusive C-CH3. Additionally, the use of C-CH2OH provides a structural water analogue to probe structural hydration effects in the minor groove. Nucleoside building blocks of the various analogues will be prepared as phosphoramidites for incorporation into DNA.
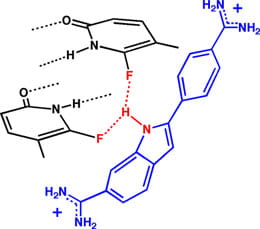
The focus of these studies is three-fold: (i) Structural Studies. Minor groove functional groups and/or the water structure or sequestered metal ions appear to contribute both to duplex stability and to duplex conformation. These effects can be probed by replacing the minor groove O2-carbonyls with derivatives that do not function as hydrogen bond acceptors (see above, right). The structural water analogue (dA tethering a covalently bound –OH) will be used in some studies. Sequences containing varying analogues nucleosides in varying amounts will be probed for stability and conformation effects as well as their ability to impact the A-tract DNA curvature effects. (ii) Ligand/Protein Binding. We will continue our studies of the importance of minor groove functionality on the binding by minor groove ligands (see above, left). Additionally, the important transcriptional pre-initiation complex – TBP-TATA box complex – one that is critically important for eukaryotic transcription will be studied. The complex itself is unusual in that the protein interacts with the A/T rich recognition sequence by binding solely through the minor groove. The minor groove base analogues will be used both to examine the importance of functional group interactions in the minor groove, as well as to determine the energetic cost of burying a hydrophilic functional group at the hydrophobic interface between the protein and the nucleic acid. (iii) Polymerase Activity. DNA polymerases and/or reverse transcriptases from bacterial, mammalian and viral origins will be studied using promoter analogues lacking specfic functional groups. We hope to define what DNA characteristics at promoter sites are “recognized” by polymerases.
The studies described in this project will help to further the understanding of DNA structure as well as sequence-specific protein-nucleic acid and ligand-nucleic acid interactions by elucidating the location and relative importance of functional group interactions that contribute to overall specificity and affinity in such complexes.
Modified Nucleosides to Probe Polymerase Activity as a Route to New Antivirals
Viral diseases continue to offer one of the great challenges to world health. Developing a better understanding of the differences in the ways that human and viral polymerases use common dNTPs offers the possibility of a new strategy for antiviral therapy. Using that knowledge, compounds could then be designed solely as viral substrates in order to terminate viral replication in the presence of host replication. Preliminary results suggest that minor groove functional groups may hold the key to such discrimination. We develop this project area as follows:
(i) Synthesis: Prepare a set of analogue nucleoside phosphoramidites and triphosphates that lack critical “minor groove functional groups” (e.g., the pyrimidine O2-carbonyl or the purine N3-nitrogen). The so-called native minor groove functional group will initially be replaced by hydrogen, but also with other functionality (-F and -CH3) that better mimic the “shape” of the native residue. Analogues will be used to probe reverse transcriptases as well as DNA and RNA polymerases.
The common nucleoside dT vs. the 2′-deoxynucleosides with the nucleobase 3-methyl-2-pyridone (dm32P – a dT analogue lacking the O2-carbonyl but otherwise maintaining normal W-C hydrogen bonding , and two better “shape” mimics of dT (or dU) using -F (dF2P) or -CH3 (dM2P) in place of the O2-carbonyl
(ii) Analogues as Polymerase Substrates: Measure the substrate effectiveness (or inhibition effects) with various polymerases for the analogue triphosphates: these studies will include single nucleotide extensions, further elongations and detailed kinetic assays.
(iii) Analogues in the Primer and/or Template: Probe the importance of minor groove functional groups in both the template and primer strands on polymerase activity. Determine the effects of such sequences on polymerase and exonuclease activities.
(iv) Chain-Terminating Derivatives: Use the information obtained from the analogue studies to design and test related compounds for their ability to terminate viral (HIV or HBV) replication. Design and prepare neutral phosphate and phosphorothioate derivatives as pro-drugs – the latterupon incorporation by the virus result in chain termination that is not easily rectified by an exonuclease activity.
(v) Purine Steric Derivatives: Prepare two purine analogues (right) in which the minor groove functional groups are absent, and replaced with methyl groups to sterically disrupt potential contacts made by polymerases. Test these for substrate activities
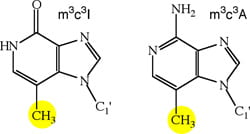
Our preliminary observations, that selective nucleobase modification can eliminate substrate activity with human polymerases but maintain activity with viral polymerases such as HIV RT, represents enzyme discrimination that should dramatically reduce the toxicity effects but maintain the antiviral effects associated with this new class of nucleoside chain terminators.
Targeting Double-Stranded DNA Using the Janus-Wedge Format
We propose to develop a fundamentally new approach to nucleic acid targeting of DNA duplexes. In the proposed recognition complexes, a third strand of DNA inserts itself between the two Watson-Crick faces of the duplex to result in a new type of DNA triplex. These triplexes are termed Janus Wedge triplexes after the Roman god Janus. Janus stood for the beginning of everything, guardian of heaven and of all gates and doors. Janus is typically pictured with two faces (below), supposedly so he could see what was coming and going. The Janus Wedge (Jw) residue in the Janus Wedge triplex, contains two hydrogen bonding faces such that after insertion into the DNA duplex, the Jw residue is hydrogen bonded to both Watson-Crick faces of the target base pair (below). The final three-stranded complex contains significantly more interstrand hydrogen bonds than does the target Watson-Crick DNA duplex, and should therefore be thermodynamically favored.
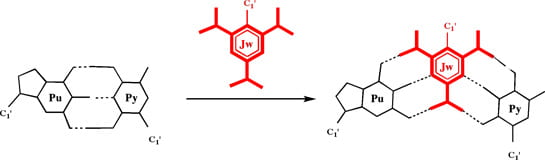
The development of the Janus Wedge concept requires a number of steps:
• (i) Prepare Jw monomers and oligomers based upon both a pyrimidine-like structure, and a purine-like structure, and study the nature and stability of Jw triplexes formed in the absence of competing Watson-Crick hydrogen bonding. Initially the triplex will feature non-complementary purine-pyrimidine base-pair targets and the Jw residue.
• (ii) Synthesize a Jw pyrimidine-like monomer and oligomers that can be used to target A-T base pairs in a DNA duplex, and upon rotation of the glycosidic bond can target also T-A base pairs. A second monomer will be prepared for recognition of the G-C/C-G base pairs.
• (iii) Use a common recognition motif and prepare four Jw purine-like monomers and corresponding oligomers designed to recognize the four common DNA base pairs, in each case forming an isomorphic Jw base triplet and generalizing this targeting format.
• (iv) Examine mechanisms for the invasion of DNA duplexes by the Jw format by using (a) Peptide nucleic acids (PNA) or other neutral backbones for the Jw strand, (b) appropriate tethered ligands to direct the Jw to its target, and (c) the process of DNA breathing to disrupt the target duplex.
The success of this project will provide a one-base to one-base-pair targeting format for the recognition of all four DNA base pairs, and thus provide a motif for generalizing the process of DNA duplex recognition using the triple helix format.
Nanoscale DNA Lattices
Supramolecular mesoscale DNA lattices (Figures 1a and 1b) formed from the hybridization of building blocks containing “hubs” to which multiple DNA “arms” are tethered, differ fundamentally from dendrimers (Figure 1c) or other aggregation assemblies since each building block for the lattice attaches to the growing supramolecular assembly in a regular geometric fashion at more than one site. In this respect such assemblies can be viewed as macromolecular versions of crystals as illustrated by example for a tetrahedral (diamond) lattice (Figure 1a) and an octahedral (cubic) lattice (Figure 1b).
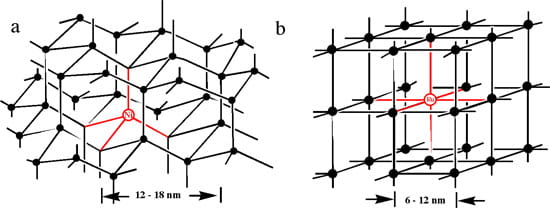
Figure 1. (a) A diamond lattice formed on the basis of a four-armed tetrahedral Ni(II) building block (red). (b) A cubic lattice formed on the basis of an octahedral six-armed Ru(II) building block (red). In both assemblies the black lines and circles represent additional DNA duplex arms and branch points, respectively.
Macromolecular lattice-like assemblies will be valuable for the study of biological processes and could provide new approaches to the controlled release of macromolecular pharmaceuticals. At least three types of properties of these ordered lattices will be studied:
(a) The DNA sequence arms: the lattices are designed such that any complementary DNA duplexes can be employed as the connecting arms. Appropriate sequence choice will permit the construction of complex lattices resulting from the binding of complementary ligands, proteins, other DNA sequences or even non-biologicals. A terminal sulfhydryl will permit the lattices to be surface bound as mono- or multilayered materials. Both assembly and binding events will be monitored by surface plasmon resonance.
(b) The metal hubs: the hubs or branch points of the described lattices are designed as metal-ligand complexes to reinforce the geometry inherent in the lattice structures. These metal centers could also function as appropriate functional sites, with the DNA arms simply present as the desired spacers. Lattices that function as light-harvesting arrays will be constructed and studied using appropriate light absorbing antennae and metal centers.
(c) The pores: these assemblies contain macropores with dimensions of tens of nanometers. The pores are defined as the spaces in between the DNA “arms” and “hubs.” The pores can increase or decrease in size, within limits, simply by varying the length of the DNA sequence arms; that not function as rulers. We will examine the porosity of the lattices as well as the ability to sequester macromolecules within the nanometer sized pores (6 – 18 nm) and similarly the rates at which nanoscale biologicals or related materials can be released from the pores.
The described DNA lattices should provide fairly rigid but biologically “friendly” structures that can be used to monitor biological events, particularly those involving DNA binding or related processes. But beyond such conventional uses, these ordered materials offer new ways in which biologicals or related bioinorganic systems can be organized into repeating three-dimensional arrays with dimensions in the nanometer or tens of nanometers range.
Alternative Carbohydrate Backbones for Information Storage Polymers (a collaborative project with Jack Szostak, HHMI, Harvard University)
The DNA/RNA carbohydrate backbone is a critical element of nucleic acids that helps to facilitate effective Watson-Crick base pairing, a crucial aspect of information transfer in biological systems. Although evolution on planet earth has resulted in nucleic acid backbones composed of D-pentofuranose sugars, this choice is by no means a limiting structure for polymers capable of information storage and transfer. Simpler backbones might be evolutionary progenitors of the proposed “RNA world”. These progenitors would have carried out receptor-ligand binding as well as catalytic reactions in an early stage in the evolution of life.
This project follows these steps:
(i) Synthesize nucleoside triphosphate building blocks and corresponding fully protected phosphoramidites, containing the T, C, A (or D) and G heterocycles, but having simpler carbohydrate backbones. The targeted backbones include the α-L-threofuranose (TNA) as well as two acyclic backbones, one based upon a glycerol methylene chain (GmNA) and the second based upon simple glycerol (GNA).
(ii) Prepare oligonucleotides of varying sizes composed of the above building blocks. Characterize duplex stability with homo-oligomers and hetero-oligomers containing native (DNA and RNA) as well as modified carbohydrate backbones. Characterize enzymatic reactions using a variety of polymerases and both native and modified NTPs with both native and analogue templates. Determine which backbones are capable of information transfer with modern polymerases.
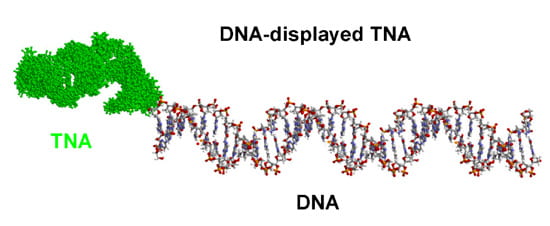
(iii) Select for functional molecules, including receptors (aptamers) and catalysts, composed of TNA, GNA or GmNA. Transcribed TNA/GNA/GmNA can be covalently linked to its DNA template (see above), functional selections can be performed for these molecules, and the successful genotypes can be rescued by PCR amplification of the attached DNA (analogous to the selection of functional peptides and proteins by mRNA-display). Characterize binding or catalytic properties of the new genotypes.
(iv) Determine whether the three alternative polymers and the analogue NTPs are substrates or templates for any available RNA polymerases. RNA can also function as an intermediate in the search for functional molecules.