Research Direction 1. Method development for quantitative detection and site-selective modulation of RNA chemical modifications in human cells.
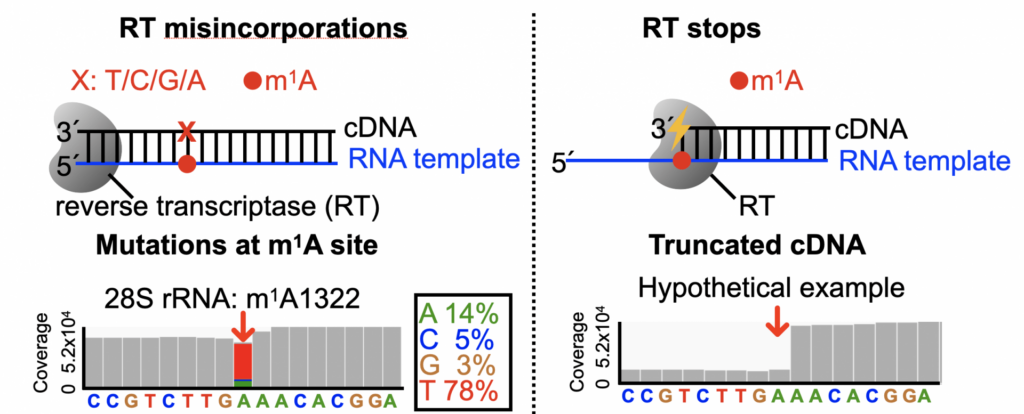
RNA chemical modifications remain challenging to be detected precisely and quantitatively due to their diverse chemical nature, and the highly transient and dynamic nature of their carrier – endogenous RNA. Recent advances in the next-generation sequencing (NGS) technologies enable the transcriptome-wide mapping of RNA chemical modifications at base-resolution for only a few types of modifications. We aim to expand the detection toolkit for RNA modifications via directed evolution of reverse transcriptases, modification-selective chemical reactions, and development of indispensable bioinformatics data analysis pipelines. Meanwhile, we are interested in developing methods for manipulating chemical modifications in a site-selective manner. We will apply the state-of-the-art tools in the investigation of biological functions of RNA chemical modifications in both coding and non-coding RNAs.
Research Direction 2. Molecular Recognitions between RNA Chemical Modifications and Reader Proteins.
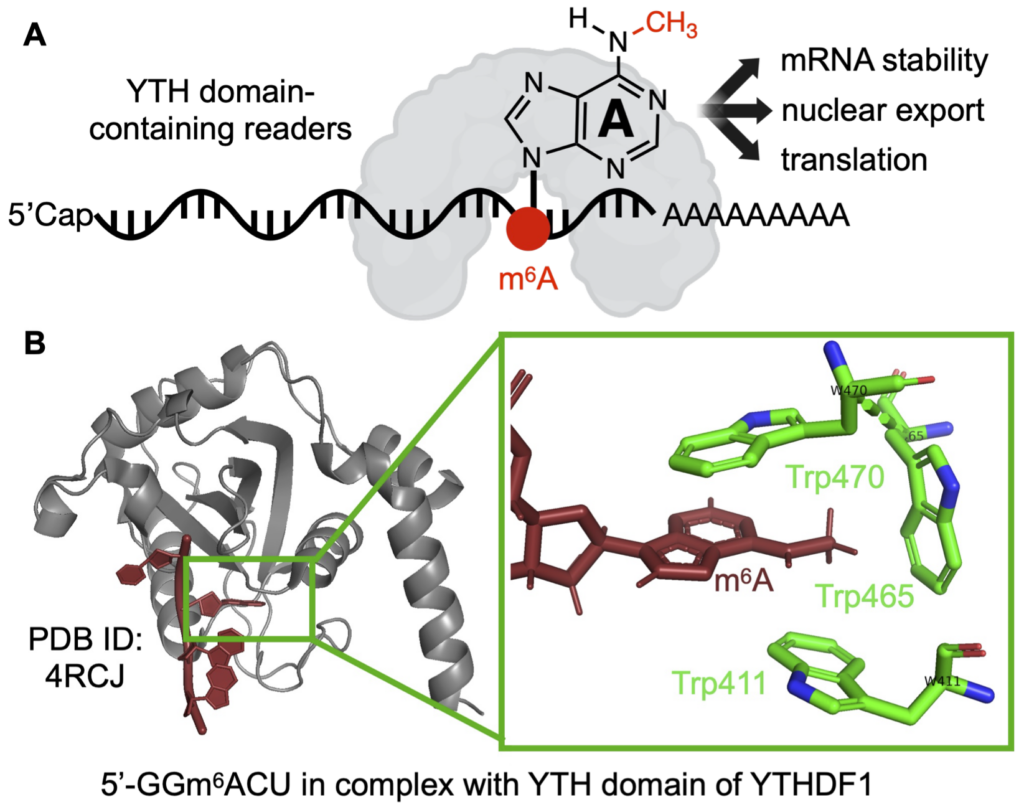
Molecular recognition between chemical modified RNA and reader proteins is critical for modifications exerting biological functions.
The recognition of m6A by the YTH-domain containing readers is achieved by a Trp-rich hydrophobic binding pocket wrapping around the N6-methyl group.
We are interested in developing tools for modulating reader recognition, and understanding the modification selectivity and sequence specificity of reader recognition.
Research Direction 3. Development of polymerases for the detection of short tandem repeats (STRs) DNA and RNA.
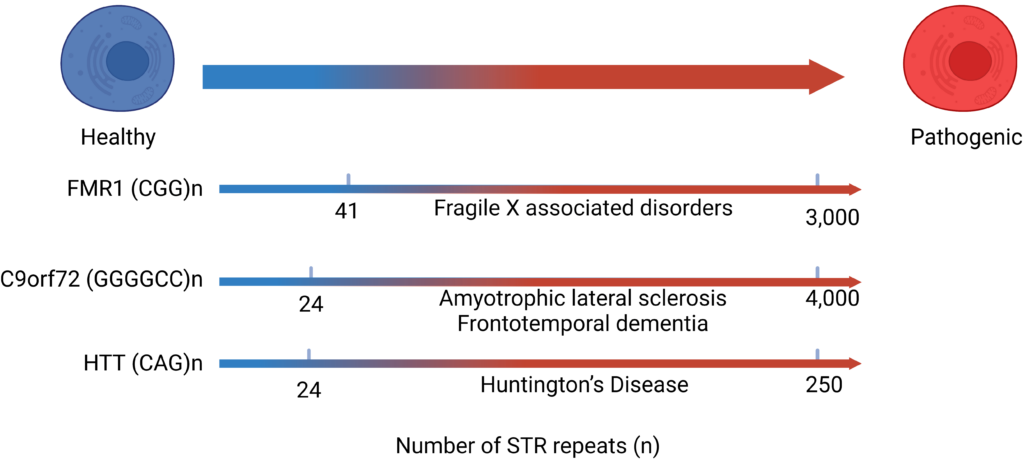
Short tandem repeats (STR) represent characteristic sequences consisting of a repeated pattern of 1-6 nucleotides, occupying around 3% of the human genome. Mutations such as repeat expansion of STR sequences often cause dysregulations in cellular functions and lead to diseases, particularly neurological disorders. Despite the central roles of STRs, investigations of functions of STR DNAs and RNAs remain largely hampered by lack of tools for effective amplifications. We aim to develop assays and tools that allows robust detection of disease-associated GC-rich STRs sequences.