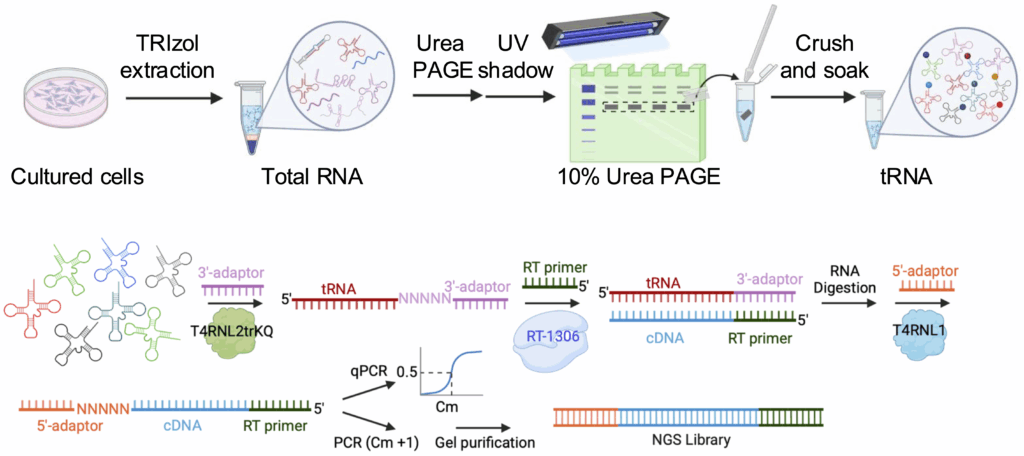
20. Tepe, M.1; Qiu, W.1; Mimouni, K.; Zhou, H.* (2025) Preparation of enzymes and libraries for MapID-tRNA-seq to identify chemical modifications in human tRNAs. Methods Enzymol. 725, 177-221. DOI: 10.1016/bs.mie.2025.10.004
19. Wang, C.; Zhou, H*. (2025) Directed evolution of an m6A eraser for site-selective epitranscriptome editing. PREPRINT available at Research Square https://doi.org/10.21203/rs.3.rs-7958216/v1.
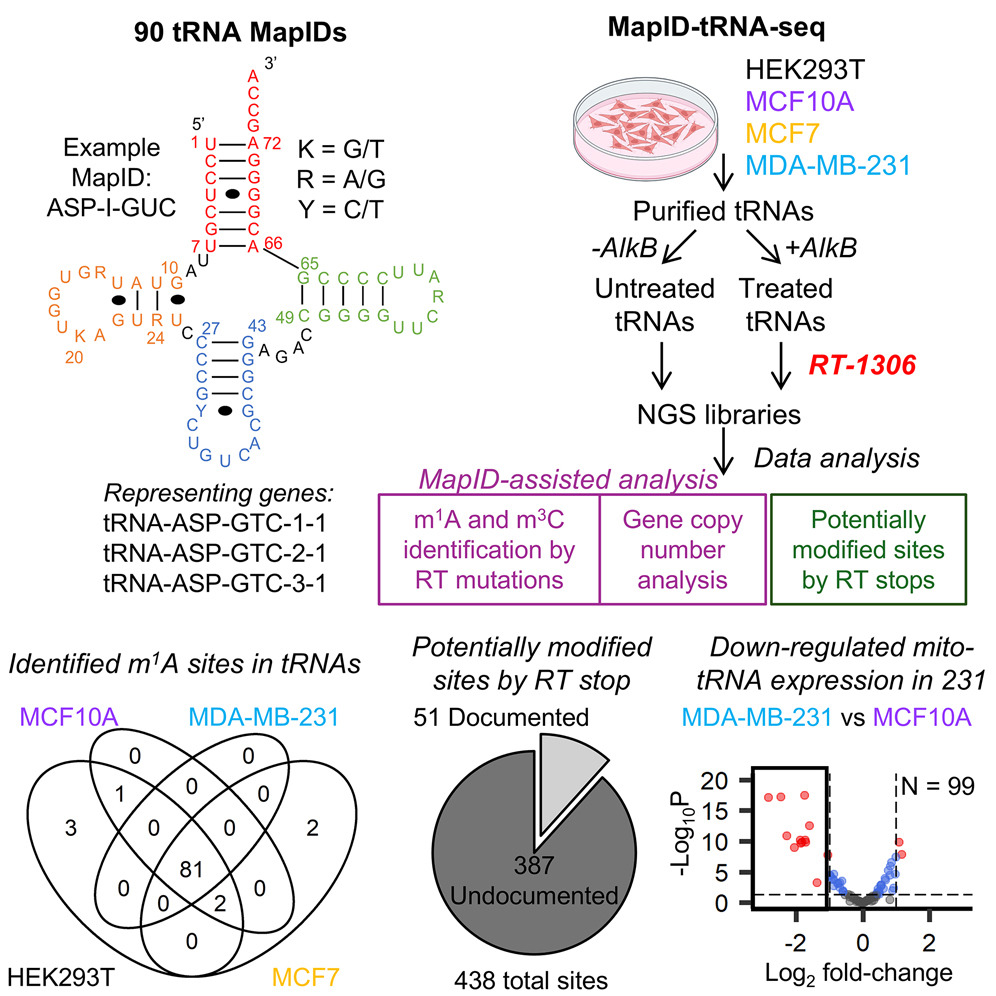
18. Tepe, M.; Chen, Y.; Carso, A.; Zhou, H.* (2025) MapID-based Quantitative Mapping of Chemical Modifications and Expression of Human Transfer RNA. Cell Chem. Biol. 5, 752-766. DOI: 10.1016/j.chembiol.2025.04.003
Check more about our perspectives:
Tepe, M.; Zhou,H. (2025) Meet the authors: Mitchel L. Tepe and Huiqing Zhou. Cell Chem. Biol. 5, 649-650. DOI: 10.1016/j.chembiol.2025.04.009
17. Qiu, W.; Hazard, C.; Li, Y.; Jin, P.; Zhou, H*. (2025) High-sensitivity Fluorescence-based Detection of Reverse Transcriptase Read-through of GC-rich Short Tandem Repeat RNA. Anal. Chem. 97, 4111-4119. https://pubs.acs.org/doi/full/10.1021/acs.analchem.4c06236

16. He, Z.; Qiu, W.; Zhou, H*. (2025) Promoted Read-through and Mutation Against Pseudouridine-CMC by an Evolved Reverse Transcriptase. Commun. Biol. 8. 40. DOI: https://doi.org/10.1038/s42003-025-07467-4 (BioRxiv. 2024.07.03.601893)

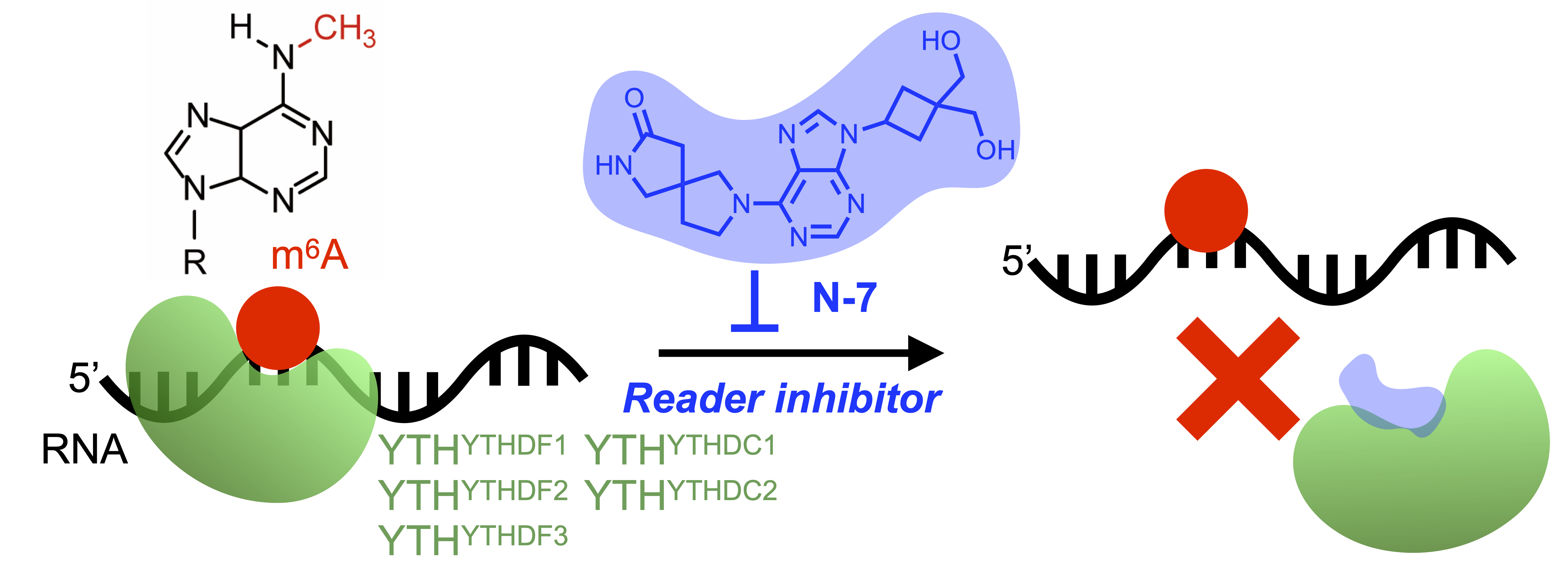
15. Wang, C.; Zhou, H*. (2024) Discovery of a New Inhibitor for the YTH Domain-containing m6A RNA Readers. RSC Chem. Biol. 5. 914-923. DOI: https://doi.org/10.1039/D4CB00105B
14. Chen, L.; Zhang, L.; Ye, C.; Zhou, H.;Liu, B.; Gao, B.; Deng, Z.; Zhao C.*; He, C.*; Dickinson, B.* (2023) Nm-Mut-seq: a base-resolution quantitative method for mapping transcriptome-wide 2′-O-methylation. Cell Research. 33. 727-730. DOI: 10.1038/s41422-023-00836-w
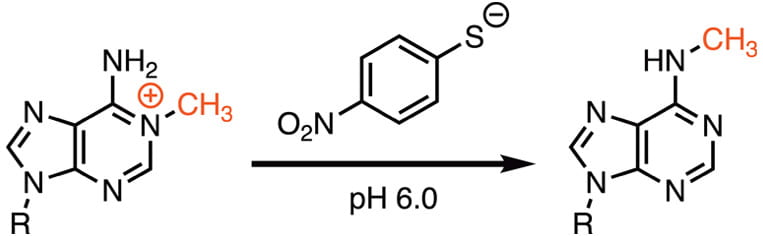
13. Liu, H.; Zeng, T.; He, C.; Rawal, V.H.; Zhou, H*.; Dickinson, B.C*. (2022) Development of Mild Chemical Catalysis Conditions for m1A-to-m6A Rearrangement on RNA. ACS Chem. Biol. 17, 1334-1342. DOI: 10.1021/acschembio.2c00178
12. Rangadurai, A.; Shi, H.; Xu, Y.; Liu, B.; Assi, H. A.; Boom, J.D.; Zhou, H.; Kimsey, I. J.; Al-Hashimi, H.M. (2022) Measuring thermodynamic preferences to form non-native conformations in nucleic acids using melting experiments reveals a rich sequence-specific DNA conformational landscape. Proceedings of the National Academy of Sciences. 119, e2112496119. DOI: 10.1073/pnas.2112496119
11. Shafik, A.M., Zhou H., Lim J., Dickinson, B.C., Jin P. (2021) Dysregulated mitochondrial and cytosolic tRNA m1A methylation in Alzheimer’s disease. Hum. Mol. Genet. ddab357.
Prior to 2020
10. Zhou, H.; Rauch, S.; Dai, Q.; Cui, X.; Zhang, Z.; Nachtergaele, S.; Sepich, C.; He, C.; Dickinson, B.C. (2019) Evolution of A Reverse Transcriptase to Map N1-Methyladenosine in Human mRNA. Nature Methods. 16, 1281-1288.
9. Rauch, S.; He E.; Scienc M.; Zhou, H.; Zhang, Z.; Dickinson, B.C. (2019) Programmable RNA-Guided RNA Effector Proteins Built from Human Parts. Cell. 178, 8-9.
8. Zhou, H.; Sathyamoorthy, B.; Stelling, A.; Xu, Y.; Xue, Y.; Pigli, Y. Z.; Case, D.; Rice, P. A.; Al-Hashimi, H. M. (2019) Resolving crystallographic ambiguity in Watson-Crick versus Hoogsteen base-pairing in a DNA-protein complex using NMR and site-specifically 13C/15N labeled DNA. Biochemistry, 58, 1963-1974.
7. Rangadurai, A.; Zhou, H.; Merriman, D.; Meiser, N.; Liu, B.; Shi, H.; Szymanski, E.; Al-Hashimi, H.M. (2018) Why are Hoogsteen base pairs energetically disfavored in A-RNA compared to B-DNA? Nucleic Acids Res., 46, 11099-11114.
6. Sathyamoorthy, B.; Shi, H.; Zhou, H.; Xue, Y; Rangadurai A.; Merriman D. K.; Al-Hashimi, H. M. (2017) Insights into Watson-Crick/Hoogsteen Breathing Dynamics and Damage Repair from the Solution Structure and Dynamic Ensemble of DNA Duplexes containing m1A. Nucleic Acids Res., 45, 5586-5601.
5. Stelling, A.; Xu, Y.; Zhou, H.; Choi, S.; Clay, M.; Merriman, D.; Al-Hashimi, H. M. (2017) Robust IR Based Detection of Stable and Fractionally Populated G‐C+ and A‐T Hoogsteen Base Pairs in Duplex DNA. FEBS Letters, 591, 1770-1784.
4. Zhou, H.; Kimsey, I. J.; Nikolova, E. N.; Sathyamoorthy, B.;Grazioli, G.; McSally, J.;Bai, T.; Wunderlich, C. H.; Kreutz,C.; Andricioaei, I.;Al-Hashimi, H. M. (2016) m1A and m1G disrupt A-RNA structure through the intrinsic instability of Hoogsteen base pairs. Nat. Struct. Mol. Biol., 23, 803-810.
3. Zhou, H.; Hintze B.J.; Kimsey, I. J.; Sathyamoorthy, B.; Yang S.; Richardson, J.S.; Al-Hashimi, H. M. (2015) New insights into Hoogsteen base pairs in DNA duplexes from a structure-based survey. Nucleic Acids Res., 43, 3420-3433.
2. Goh, G.B.; Hulbert B.S.; Zhou, H.; Brooks. C.L. 3rd. (2014) Constant pH molecular dynamics of proteins in explicit solvent with proton tautomerism. Proteins, 82, 1319-1331.
1. Nikolova, E.N.; Zhou, H.; Gottardo F.L.; Alvey, H. S.; Kimsey, I. J.; Al-Hashimi, H. M. (2013) A Historical Account of Hoogsteen Base-Pairs in Duplex DNA. Biopolymers,99, 955-968.